Category:Transcription (genetics)
Jump to navigation
Jump to search
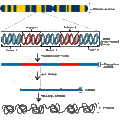
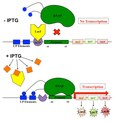
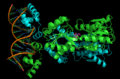
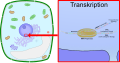
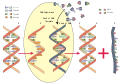
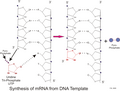
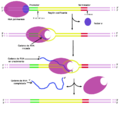
English: Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes. During transcription, a DNA sequence is read by RNA polymerase, which produces a complementary, antiparallel RNA strand. As opposed to DNA replication, transcription results in an RNA complement that includes uracil (U) in all instances where thymine (T) would have occurred in a DNA complement.
biosynthesis of RNA carried out on a template of DNA | |||||
| Upload media | |||||
| Subclass of | |||||
|---|---|---|---|---|---|
| Part of |
| ||||
| Different from | |||||
| |||||
Subcategories
This category has the following 13 subcategories, out of 13 total.
B
- Beta-catenin signaling (10 F)
G
M
R
S
- Start codon (7 F)
T
- Transcriptional activation (20 F)
- Transcriptome (26 F)
V
Media in category "Transcription (genetics)"
The following 200 files are in this category, out of 269 total.
(previous page) (next page)-
"Flowerbed" table of genetic code.png 4,716 × 3,346; 1.7 MB
-
1-s2.0-S1874939913000436-gr1 lrg.jpg 3,160 × 2,991; 654 KB
-
1a02.png 500 × 500; 147 KB
-
1gtw.png 600 × 600; 135 KB
-
5' cap labeled.svg 1,745 × 1,388; 209 KB
-
5' cap structure.png 512 × 458; 18 KB
-
5' cap structure.svg 442 × 338; 51 KB
-
5'-Cap structure.png 1,394 × 734; 60 KB
-
5primeUTR PictorialDefinition.png 648 × 360; 3 KB
-
Abortive cycling.png 400 × 228; 37 KB
-
AlternativeSplicing uk.png 1,280 × 1,024; 66 KB
-
AlternativeSplicing.png 1,280 × 1,024; 74 KB
-
Antisense DNA oligonucleotide.png 2,750 × 1,424; 1.14 MB
-
Ayuste-alternativo.JPG 1,280 × 1,024; 103 KB
-
Basic diagram of locations of gene expression ku.jpg 1,626 × 1,080; 174 KB
-
Basic diagram of locations of gene expression.jpg 939 × 711; 127 KB
-
BHLH i seqüència consens.jpg 1,013 × 632; 100 KB
-
BHLH.jpg 583 × 439; 50 KB
-
Biology-02-00064-g001.png 1,105 × 1,046; 387 KB
-
Blocking Splicing.png 768 × 576; 12 KB
-
Cap structure.svg 640 × 320; 46 KB
-
Cap Structure.svg 355 × 210; 63 KB
-
Centralny dogmat biologii molekularnej.png 1,300 × 966; 46 KB
-
Chinese-braille.ogg 33 min 50 s; 12.62 MB
-
Class I virus (dsDNA) RNA synthesis.svg 786 × 69; 17 KB
-
Class II virus (ssDNA) RNA synthesis.svg 786 × 106; 47 KB
-
Class III virus (dsRNA) RNA synthesis.svg 786 × 59; 20 KB
-
Class IV virus (ssRNA +) RNA synthesis.svg 786 × 59; 34 KB
-
Class V virus (ssRNA -) RNA synthesis.svg 786 × 59; 20 KB
-
Class VI virus (ssRNA-RT) RNA synthesis.svg 786 × 69; 51 KB
-
Class VII virus (dsDNA-RT) RNA synthesis.svg 788 × 137; 55 KB
-
Comparison typical enhancer and super enhancer.jpg 2,550 × 3,300; 1.67 MB
-
Core promoter elements.jpg 1,280 × 535; 88 KB
-
Core promoter elements.svg 800 × 335; 57 KB
-
Crystal structure of the MafA homodimer.png 500 × 500; 86 KB
-
CUT&RUN Protocol.tif 3,163 × 3,904; 3.01 MB
-
Diagram showing the difference between DNA, RNA, and proteins.png 1,745 × 877; 15 KB
-
DNA ranscription ar.png 2,000 × 1,213; 326 KB
-
DNA to mRNA processing.jpg 1,085 × 370; 37 KB
-
DNA to mRNA processing.svg 512 × 175; 52 KB
-
DNA Transcription cyle.jpg 3,193 × 1,949; 592 KB
-
DNA transcription ku.png 1,534 × 877; 433 KB
-
DNA transcription UKR.png 884 × 536; 125 KB
-
DNA transcription.jpg 553 × 231; 56 KB
-
DNA transcription.png 1,000 × 552; 68 KB
-
DNA-RNA D-loops, greatly exaggerated.jpg 2,562 × 2,731; 243 KB
-
Dogme-central-BioMol ar.svg 336 × 122; 73 KB
-
Dogme-central-BioMol.svg 336 × 122; 25 KB
-
Dp event.png 303 × 73; 552 bytes
-
Editosome.PNG 721 × 385; 67 KB
-
Epigenetic priming model.png 4,096 × 4,096; 1.75 MB
-
EQTL.jpg 614 × 336; 24 KB
-
Estrutura Cap 5'.png 750 × 674; 101 KB
-
Estrutura de regulação da Pol III.jpg 355 × 267; 18 KB
-
Etls-2019-0024c.01.png 1,822 × 1,277; 311 KB
-
Eukaryotic RNA Polymerase II rotating.gif 349 × 237; 6.01 MB
-
Eukaryotic Transcription.gif 450 × 450; 32 KB
-
Eukaryotic Transcription.png 914 × 676; 259 KB
-
Eukaryotic Transcription.svg 450 × 450; 156 KB
-
Eukaryotic transcripton bubble with and without RNA Polymerase II.png 2,218 × 604; 638 KB
-
Extended Central Dogma with Enzymes.jpg 762 × 389; 124 KB
-
Fbioe-09-718753-g002.jpg 673 × 350; 24 KB
-
Fgene-10-00006-g001.jpg 1,000 × 738; 296 KB
-
Fgene-10-00006-g002.jpg 1,000 × 803; 402 KB
-
Figura 2.jpg 334 × 251; 17 KB
-
Figura 3.1.jpg 417 × 270; 21 KB
-
Figura 4.1.jpg 571 × 429; 46 KB
-
Figura-Trans-splicing.png 537 × 705; 7 KB
-
Fimmu-09-01581-g003.jpg 1,192 × 690; 394 KB
-
Fimmu-09-01581-g004.jpg 1,192 × 610; 331 KB
-
Five prime cap structure.svg 474 × 302; 84 KB
-
Gapmer mechanism of action updated.png 660 × 647; 98 KB
-
Gapmer mechanism of action.png 660 × 644; 82 KB
-
Gene promoters according to T1D, IDM and T2D.png 2,205 × 1,469; 790 KB
-
Gene structure 2 annotated.svg 1,199 × 657; 675 KB
-
Gene structure eukaryote 2 annotated (hyperlinked).svg 1,190 × 640; 158 KB
-
Gene structure eukaryote 2 annotated ku.svg 1,190 × 640; 25 KB
-
Gene structure eukaryote 2 annotated.svg 1,190 × 640; 15 KB
-
Gene structure prokaryote 2 annotated (hyperlinked).svg 1,190 × 586; 143 KB
-
Gene structure prokaryote 2 annotated ku.svg 1,190 × 586; 140 KB
-
Gene structure prokaryote 2 annotated.svg 1,190 × 586; 136 KB
-
Genetik-Translation-Transkription-leer.svg 512 × 729; 5.08 MB
-
Genetik-Translation-Transkription.svg 512 × 827; 6.32 MB
-
Gràfic Yubi v5.jpg 2,305 × 1,349; 1.6 MB
-
Histone tails set for transcriptional activation.jpg 2,346 × 2,082; 244 KB
-
Histone tails set for transcriptional repression.jpg 2,315 × 2,048; 583 KB
-
Image for Wiki 1.jpg 1,200 × 1,002; 145 KB
-
Image for Wiki 2.jpg 487 × 327; 35 KB
-
Informazioaren irakurketa.tif 744 × 363; 137 KB
-
Insertion uk.png 628 × 395; 101 KB
-
Intein splicing dogma.png 1,027 × 742; 150 KB
-
Intrinsic Termination Structure.png 526 × 546; 220 KB
-
Journal.pone.0083796.g007.png 1,759 × 1,162; 1.12 MB
-
Journal.ppat.1000772.g001.tif 2,765 × 4,147; 1.3 MB
-
Label RNA pol II.png 300 × 441; 137 KB
-
Lac operon.pdf 1,089 × 1,131; 277 KB
-
Lac-Repressor.jpg 1,920 × 1,920; 712 KB
-
LacIOperatorComplex.png 1,014 × 664; 406 KB
-
MechanismIntrinsicTerm.jpg 1,280 × 506; 120 KB
-
MRNA (editorin versio).svg 1,743 × 1,766; 135 KB
-
MRNA ar.svg 1,743 × 1,766; 1.21 MB
-
MRNA kn.png 1,743 × 1,767; 713 KB
-
MRNA ku.svg 1,743 × 1,766; 158 KB
-
Mrna trans2.png 720 × 697; 94 KB
-
MRNA-fr.svg 1,817 × 1,760; 1.53 MB
-
MRNA-id.svg 1,743 × 1,766; 886 KB
-
MRNA-interaction nl.svg 448 × 453; 284 KB
-
MRNA-interaction-idn.png 448 × 453; 55 KB
-
MRNA-interaction.png 448 × 453; 47 KB
-
MRNA-interaction.svg 600 × 600; 66 KB
-
MRNA.svg 1,743 × 1,766; 1.23 MB
-
MyoD recruitment.png 26,750 × 5,000; 2.16 MB
-
Nantitermination1.jpg 717 × 388; 50 KB
-
Nantitermination2.jpg 711 × 447; 75 KB
-
-
-
-
-
Nucleosome at enhancer with H3K122 acetylated.jpg 2,315 × 2,048; 623 KB
-
-
NUP-1-Is-a-Large-Coiled-Coil-Nucleoskeletal-Protein-in-Trypanosomes-with-Lamin-Like-Functions-pbio.1001287.s007.ogv 9.0 s, 2,048 × 2,048; 163 KB
-
-
-
-
-
Pdr1p and Transcripton Schematic.png 1,039 × 503; 77 KB
-
Peptide syn-el.svg 720 × 460; 68 KB
-
Pflanzenzelle mit Proteinbiosynthese.svg 512 × 360; 4.67 MB
-
Pflanzenzelle-Transkription.svg 512 × 266; 5.79 MB
-
Polyadenylation.png 1,024 × 1,877; 245 KB
-
Preinitiation complex ru.svg 330 × 95; 38 KB
-
Preinitiation complex.png 330 × 100; 13 KB
-
Proceso de transcripción.jpg 2,007 × 924; 56 KB
-
Process of transcription (13080846733).jpg 842 × 595; 124 KB
-
Prokariotic terminators-gl.svg 430 × 664; 20 KB
-
Prokaryotic terminators-en.svg 430 × 664; 20 KB
-
Proposed Stdb1-MyoD pathway.png 14,663 × 4,557; 738 KB
-
-
-
-
-
Proteines.png 538 × 348; 15 KB
-
PV SSM.JPG 960 × 720; 69 KB
-
Regualação da transcrição pela pol III.jpg 417 × 314; 18 KB
-
Regulation of Lactose Metabolism in Prokaryotes.svg 512 × 384; 134 KB
-
Regulation of transcription in mammals.jpg 2,524 × 1,372; 227 KB
-
Regulation of transcription in metazoans (animals).jpg 2,524 × 1,604; 548 KB
-
Remodeladores de la cromatina y regulación de la transcripción.png 1,206 × 844; 222 KB
-
Repressors modulate transcription output..jpg 552 × 346; 51 KB
-
RetroTranscription.jpg 640 × 480; 46 KB
-
Rho And Intrinsic Termination photo.jpg 1,805 × 1,293; 139 KB
-
RibosomaleTranskriptionsEinheit (cropped).jpg 608 × 498; 84 KB
-
RibosomaleTranskriptionsEinheit.jpg 1,039 × 1,250; 729 KB
-
RNA polymerase activity (transcription).gif 1,200 × 765; 1.56 MB
-
RNA polymerase transcribing template strand DNA.png 1,648 × 956; 173 KB
-
RNA primer.png 720 × 380; 63 KB
-
Rna syn.png 720 × 536; 133 KB
-
Rna syn.svg 725 × 505; 32 KB
-
Rna syn2.png 648 × 493; 65 KB
-
RNA transcription and template switch.png 1,960 × 1,770; 928 KB
-
RNA-transcription-processing-pt-br.png 2,752 × 1,644; 543 KB
-
RNA-transskribado.svg 1,859 × 1,884; 1.41 MB
-
RNAP TEC small.jpg 302 × 292; 86 KB
-
Rnapol.png 984 × 839; 713 KB
-
RNAtranscription.png 1,024 × 1,024; 57 KB
-
Schematischer Ablauf der DNA-Transkription.png 763 × 438; 77 KB
-
Simple transcription elongation1 NL.svg 720 × 130; 41 KB
-
Simple transcription elongation1-fr.svg 721 × 129; 37 KB
-
Simple transcription elongation1.svg 721 × 129; 36 KB
-
Simple transcription elongation2-fr.svg 769 × 138; 41 KB
-
Simple transcription initiation1-fr.svg 721 × 223; 28 KB
-
Simple transcription initiation1.svg 721 × 223; 28 KB
-
Simple transcription termination1-fr.svg 721 × 210; 38 KB
-
Simple transcription termination1.svg 721 × 210; 37 KB
-
Simplified domains of DNMT3A isoforms.jpg 1,835 × 1,213; 312 KB
-
-
SmeT and SmeDEF transcription start sites.jpg 1,225 × 664; 232 KB
-
SMIM19 Transcript Variants 1-4.png 1,872 × 394; 119 KB
-
SMIM19 Transcript Variants.png 2,294 × 1,184; 367 KB
-
-
-
-
-
-
Spatial-and-Topological-Organization-of-DNA-Chains-Induced-by-Gene-Co-localization-pcbi.1000678.s007.ogv 7.5 s, 756 × 580; 6.15 MB
-
Stem-loop.svg 336 × 229; 11 KB
-
Stochastic-mRNA-Synthesis-in-Mammalian-Cells-pbio.0040309.sv001.ogv 16 s, 789 × 612; 4.31 MB
-
Stochastic-mRNA-Synthesis-in-Mammalian-Cells-pbio.0040309.sv002.ogv 11 s, 821 × 628; 2.53 MB
-
Stochasticity-and-the-Molecular-Mechanisms-of-Induced-Pluripotency-pone.0003086.s002.ogv 10 s, 404 × 202; 130 KB
-
Stochasticity-and-the-Molecular-Mechanisms-of-Induced-Pluripotency-pone.0003086.s003.ogv 10 s, 404 × 202; 132 KB
-
Stochasticity-and-the-Molecular-Mechanisms-of-Induced-Pluripotency-pone.0003086.s004.ogv 8.0 s, 404 × 202; 112 KB
-
Stochasticity-and-the-Molecular-Mechanisms-of-Induced-Pluripotency-pone.0003086.s005.ogv 13 s, 404 × 202; 150 KB
-
Structural-Basis-of-Gate-DNA-Breakage-and-Resealing-by-Type-II-Topoisomerases-pone.0011338.s005.ogv 31 s, 800 × 750; 7.26 MB