Category:Molecular dynamics simulation
Jump to navigation
Jump to search
method of computer simulation of molecular interaction | |||||
| Upload media | |||||
| Instance of | |||||
|---|---|---|---|---|---|
| Subclass of |
| ||||
| |||||
Subcategories
This category has the following 4 subcategories, out of 4 total.
Media in category "Molecular dynamics simulation"
The following 200 files are in this category, out of 302 total.
(previous page) (next page)-
-
A-Thermodynamic-Model-of-Microtubule-Assembly-and-Disassembly-pone.0006378.s001.ogv 40 s, 800 × 192; 832 KB
-
A-Thermodynamic-Model-of-Microtubule-Assembly-and-Disassembly-pone.0006378.s002.ogv 38 s, 800 × 192; 1.93 MB
-
Aba Graphical abstract.jpg 1,680 × 886; 320 KB
-
ACS OMEGA.png 750 × 369; 250 KB
-
-
An-electrostatic-mechanism-for-Ca2+-mediated-regulation-of-gap-junction-channels-ncomms9770-s4.ogv 39 s, 752 × 1,056; 58.48 MB
-
An-electrostatic-mechanism-for-Ca2+-mediated-regulation-of-gap-junction-channels-ncomms9770-s7.ogv 40 s, 752 × 1,056; 63.66 MB
-
-
-
An-Introduction-to-Biomolecular-Graphics-pcbi.1000918.s007.ogv 2 min 2 s, 1,320 × 788; 14.4 MB
-
Anton supercomputer.jpg 1,599 × 1,059; 265 KB
-
Atomic nucleus simulated on a quantum computer - 28434461768.jpg 1,280 × 720; 1.3 MB
-
Brownian-dynamics-simulation-of-analytical-ultracentrifugation-experiments-2046-1682-4-6-S1.ogv 17 s, 576 × 240; 14.54 MB
-
Brownian-dynamics-simulation-of-analytical-ultracentrifugation-experiments-2046-1682-4-6-S2.ogv 17 s, 592 × 240; 15.14 MB
-
Calcium-Induced-Regulation-of-Skeletal-Troponin----Computational-Insights-from-Molecular-Dynamics-pone.0058313.s005.ogv 28 s, 1,680 × 1,032; 10.14 MB
-
-
-
Calcium-Signals-Driven-by-Single-Channel-Noise-pcbi.1000870.s002.ogv 6.4 s, 260 × 260; 882 KB
-
Catch-bond-mechanism-of-the-bacterial-adhesin-FimH-ncomms10738-s2.ogv 1 min 0 s, 1,136 × 804; 3.99 MB
-
Catch-bond-mechanism-of-the-bacterial-adhesin-FimH-ncomms10738-s3.ogv 1 min 0 s, 1,136 × 804; 2.78 MB
-
CDK2-Selective inhibitor.png 2,224 × 1,512; 1.63 MB
-
CellLists Ghosts.png 453 × 463; 8 KB
-
CellLists.png 353 × 665; 6 KB
-
-
-
Chk1 Inhibitor.jpg 2,213 × 869; 551 KB
-
-
-
CMV Genomics.png 3,812 × 2,900; 3.42 MB
-
Computational microscopy focusing on the drug binding to GPCR.webm 1 min 30 s, 1,920 × 1,080; 110.72 MB
-
Conformation-and-dynamics-of-the-ligand-shell-of-a-water-soluble-Au102-nanoparticle-ncomms10401-s2.ogv 50 s, 854 × 480; 29.45 MB
-
Conformation-and-dynamics-of-the-ligand-shell-of-a-water-soluble-Au102-nanoparticle-ncomms10401-s3.ogv 50 s, 854 × 480; 28.24 MB
-
Conformational-Dynamics-of-Dry-Lamellar-Crystals-of-Sugar-Based-Lipids-An-Atomistic-Simulation-Study-pone.0101110.s009.ogv 2 min 13 s, 320 × 240; 57.99 MB
-
Conformational-Dynamics-of-Dry-Lamellar-Crystals-of-Sugar-Based-Lipids-An-Atomistic-Simulation-Study-pone.0101110.s010.ogv 1 min 3 s, 640 × 480; 31.99 MB
-
Conformational-Sampling-and-Nucleotide-Dependent-Transitions-of-the-GroEL-Subunit-Probed-by-pcbi.1002004.s005.ogv 1 min 40 s, 480 × 480; 3.8 MB
-
Conservation of total energy in a molecule dynamics simulation.png 592 × 592; 33 KB
-
-
-
-
Correct-folding-of-an-α-helix-and-a-β-hairpin-using-a-polarized-2D-torsional-potential-srep10359-s2.ogv 12 min 0 s, 384 × 288; 17.92 MB
-
-
-
-
-
CP2K logo.png 800 × 800; 174 KB
-
-
-
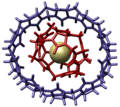
Cucurbituril gyroscope AngewChemIntEd 2002 v41 p275 hires.png 3,556 × 3,171; 2.3 MB
-
Cylindrical squalenoyl-doxorubicin nanoparticle.png 2,000 × 2,000; 3.7 MB
-
Data-of-the-molecular-dynamics-simulations-of-mutations-in-the-human-connexin46-docking-interface-mmc2.ogv 2 min 23 s, 850 × 456; 14 MB
-
Data-of-the-molecular-dynamics-simulations-of-mutations-in-the-human-connexin46-docking-interface-mmc3.ogv 2 min 5 s, 850 × 456; 10.26 MB
-
Data-of-the-molecular-dynamics-simulations-of-mutations-in-the-human-connexin46-docking-interface-mmc4.ogv 1 min 1 s, 850 × 456; 4.36 MB
-
Data-of-the-molecular-dynamics-simulations-of-mutations-in-the-human-connexin46-docking-interface-mmc5.ogv 1 min 49 s, 850 × 456; 8.09 MB
-
Data-of-the-molecular-dynamics-simulations-of-mutations-in-the-human-connexin46-docking-interface-mmc6.ogv 2 min 31 s, 850 × 456; 16.93 MB
-
-
-
-
-
-
-
Dual-Action-of-BPC194-A-Membrane-Active-Peptide-Killing-Bacterial-Cells-pone.0061541.s001.ogv 3.5 s, 589 × 444; 59 KB
-
Dual-Action-of-BPC194-A-Membrane-Active-Peptide-Killing-Bacterial-Cells-pone.0061541.s002.ogv 3.3 s, 589 × 444; 39 KB
-
Dual-Action-of-BPC194-A-Membrane-Active-Peptide-Killing-Bacterial-Cells-pone.0061541.s003.ogv 3.3 s, 589 × 444; 162 KB
-
Epilepsy.png 1,643 × 1,555; 768 KB
-
Exploring-the-Origin-of-Differential-Binding-Affinities-of-Human-Tubulin-Isotypes-αβII-αβIII-and-pone.0156048.s011.ogv 17 s, 1,840 × 1,056; 29.64 MB
-
Exploring-the-Universe-of-Protein-Structures-beyond-the-Protein-Data-Bank-pcbi.1000957.s008.ogv 0.0 s, 656 × 832; 6.59 MB
-
Filament-Depolymerization-Can-Explain-Chromosome-Pulling-during-Bacterial-Mitosis-pcbi.1002145.s009.ogv 54 s, 1,036 × 690; 20.84 MB
-
Filament-Depolymerization-Can-Explain-Chromosome-Pulling-during-Bacterial-Mitosis-pcbi.1002145.s010.ogv 22 s, 1,036 × 690; 10.5 MB
-
Filament-Depolymerization-Can-Explain-Chromosome-Pulling-during-Bacterial-Mitosis-pcbi.1002145.s011.ogv 32 s, 1,036 × 690; 11.71 MB
-
Food chemistry.png 1,194 × 886; 911 KB
-
-
-
-
-
GFPs-Mechanical-Intermediate-States-pone.0046962.s003.ogv 1 min 9 s, 852 × 480; 23.95 MB
-
Graphene oxide in liquid water.png 945 × 1,249; 458 KB
-
Graphical abstract EcR.png 646 × 331; 111 KB
-
-
-
Holec2016AtomisticMaterialsModellingP19 de.svg 512 × 353; 11 KB
-
Holec2016AtomisticMaterialsModellingP19.svg 1,184 × 817; 4 KB
-
Holec2016P40.svg 512 × 380; 10 KB
-
Homology-modeling-and-ligand-docking-of-Mitogen-activated-protein-kinase-activated-protein-kinase-5-1742-4682-10-56-S6.ogv 1 min 0 s, 320 × 240; 11.65 MB
-
Hot-Melt-Extruded-Amorphous-Solid-Dispersion-of-Posaconazole-with-Improved-Bioavailability-146781.f1.ogv 1 min 55 s, 640 × 480; 36.24 MB
-
Human tyrosyl-tRNA synthetase (interdomain interaction, TyrRS).png 3,849 × 2,913; 2.62 MB
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
Inhibition-of-Nutlin-Resistant-HDM2-Mutants-by-Stapled-Peptides-pone.0081068.s002.ogv 4.1 s, 1,440 × 1,080; 1.73 MB
-
Inhibition-of-Nutlin-Resistant-HDM2-Mutants-by-Stapled-Peptides-pone.0081068.s003.ogv 5.0 s, 1,440 × 1,080; 1.62 MB
-
Inhibition-of-Nutlin-Resistant-HDM2-Mutants-by-Stapled-Peptides-pone.0081068.s004.ogv 3.9 s, 1,440 × 1,080; 950 KB
-
Inhibition-of-Nutlin-Resistant-HDM2-Mutants-by-Stapled-Peptides-pone.0081068.s005.ogv 3.0 s, 1,440 × 1,080; 886 KB
-
Insertion-of-Short-Amino-Functionalized-Single-Walled-Carbon-Nanotubes-into-Phospholipid-Bilayer-pone.0040703.s008.ogv 1 min 0 s, 288 × 224; 6.87 MB
-
-
Insertion-of-Short-Amino-Functionalized-Single-Walled-Carbon-Nanotubes-into-Phospholipid-Bilayer-pone.0040703.s010.ogv 1 min 35 s, 288 × 224; 11.02 MB
-
Interplay-of-active-processes-modulates-tension-and-drives-phase-transition-in-self-renewing-motor-ncomms10323-s2.ogv 5.5 s, 1,000 × 1,000; 8.01 MB
-
Interplay-of-active-processes-modulates-tension-and-drives-phase-transition-in-self-renewing-motor-ncomms10323-s3.ogv 10 s, 1,000 × 1,000; 23.57 MB
-
Interplay-of-active-processes-modulates-tension-and-drives-phase-transition-in-self-renewing-motor-ncomms10323-s4.ogv 23 s, 1,000 × 1,000; 48.23 MB
-
-
Interplay-of-active-processes-modulates-tension-and-drives-phase-transition-in-self-renewing-motor-ncomms10323-s6.ogv 1 min 15 s, 512 × 512; 15.49 MB
-
-
-
-
Islands-Containing-Slowly-Hydrolyzable-GTP-Analogs-Promote-Microtubule-Rescues-pone.0030103.s003.ogv 1 min 32 s, 150 × 74; 4.81 MB
-
Jopurnal of molecular lipids.png 2,213 × 826; 1.07 MB
-
Large-Scale-Characterization-of-the-LC13-TCR-and-HLA-B8-Structural-Landscape-in-Reaction-to-172-pcbi.1003748.s008.ogv 1 min 3 s, 620 × 804; 51.21 MB
-
-
Les méthodes « hybride.png 572 × 424; 9 KB
-
Limiteperiodicite.svg 512 × 512; 112 KB
-
Linear vs. Full Poisson-Boltzmann Equation.svg 512 × 467; 212 KB
-
Linearized vs Full Potential Forms of the Poisson-Boltzmann Equation.svg 512 × 352; 108 KB
-
Main protease.jpg 3,300 × 1,723; 579 KB
-
-
-
-
-
Mapping-the-dynamics-and-nanoscale-organization-of-synaptic-adhesion-proteins-using-monomeric-ncomms10773-s4.ogv 21 s, 1,200 × 900; 31.54 MB
-
Mapping-the-dynamics-and-nanoscale-organization-of-synaptic-adhesion-proteins-using-monomeric-ncomms10773-s5.ogv 21 s, 1,158 × 897; 25.79 MB
-
Mapping-the-dynamics-and-nanoscale-organization-of-synaptic-adhesion-proteins-using-monomeric-ncomms10773-s6.ogv 6.3 s, 1,140 × 1,140; 6.88 MB
-
-
MD rotor 250K 1ns.gif 300 × 314; 6.49 MB
-
MD water.gif 480 × 270; 5.87 MB
-
MDynaMix-MGE.png 888 × 742; 115 KB
-
-
Membrane-Recognition-and-Dynamics-of-the-RNA-Degradosome-pgen.1004961.s012.ogv 17 s, 848 × 832; 16.01 MB
-
Membrane-Recognition-and-Dynamics-of-the-RNA-Degradosome-pgen.1004961.s013.ogv 6.1 s, 320 × 160; 554 KB
-
Membrane-Sculpting-by-F-BAR-Domains-Studied-by-Molecular-Dynamics-Simulations-pcbi.1002892.s003.ogv 14 s, 1,904 × 992; 7.24 MB
-
Membrane-Sculpting-by-F-BAR-Domains-Studied-by-Molecular-Dynamics-Simulations-pcbi.1002892.s004.ogv 54 s, 1,904 × 992; 13.71 MB
-
Membrane-tension-controls-the-assembly-of-curvature-generating-proteins-ncomms8219-s1.ogv 14 s, 1,920 × 614; 18.22 MB
-
-
-
Mo110s.ogv 15 s, 928 × 696; 1.57 MB
-
Modeling-a-New-Water-Channel-That-Allows-SET9-to-Dimethylate-p53-pone.0019856.s005.ogv 10 s, 656 × 624; 1.87 MB
-
Modèle simplifié de mouvement d'une molécule.png 438 × 212; 17 KB
-
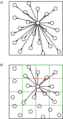
Molecular dynamics algorithm.png 1,009 × 909; 64 KB
-
Molecular Dynamics Simulation of DPPC Lipid Bilayer.webm 32 s, 630 × 982; 51.8 MB
-
Molecular gyroscope 2007 uncaged.png 2,206 × 1,284; 179 KB
-
Molecular-Mechanism-of-Binding-between-17β-Estradiol-and-DNA-mmc1.ogv 13 s, 1,280 × 720; 628 KB
-
Molecular-Mechanism-of-Cyclodextrin-Mediated-Cholesterol-Extraction-pcbi.1002020.s006.ogv 47 s, 320 × 240; 12.1 MB
-
Monitoring-dynamics-of-human-adenovirus-disassembly-induced-by-mechanical-fatigue-srep01434-s2.ogv 53 s, 720 × 540; 5.44 MB
-
Monitoring-dynamics-of-human-adenovirus-disassembly-induced-by-mechanical-fatigue-srep01434-s3.ogv 39 s, 720 × 540; 4.13 MB
-
Monitoring-dynamics-of-human-adenovirus-disassembly-induced-by-mechanical-fatigue-srep01434-s4.ogv 46 s, 720 × 540; 4.82 MB
-
Monitoring-dynamics-of-human-adenovirus-disassembly-induced-by-mechanical-fatigue-srep01434-s5.ogv 41 s, 720 × 540; 3.93 MB
-
Monitoring-dynamics-of-human-adenovirus-disassembly-induced-by-mechanical-fatigue-srep01434-s6.ogv 34 s, 720 × 540; 3.25 MB
-
Movie 7. Comparison between directed (motor) movement and diffusive movement.ogv 31 s, 774 × 582; 5.81 MB
-
Multi-Patterned-Dynamics-of-Mitochondrial-Fission-and-Fusion-in-a-Living-Cell-pone.0019879.s007.ogv 19 s, 320 × 240; 1.07 MB
-
Multi-Patterned-Dynamics-of-Mitochondrial-Fission-and-Fusion-in-a-Living-Cell-pone.0019879.s008.ogv 26 s, 640 × 480; 1.03 MB
-
-
Multi-Patterned-Dynamics-of-Mitochondrial-Fission-and-Fusion-in-a-Living-Cell-pone.0019879.s010.ogv 25 s, 640 × 480; 1.65 MB
-
-
Mussel-adhesion-is-dictated-by-time-regulated-secretion-and-molecular-conformation-of-mussel-ncomms9737-s3.ogv 1 min 25 s, 960 × 540; 14.9 MB
-
Méthode Lattice.png 735 × 429; 20 KB
-
Méthodes Particulaire.png 308 × 158; 26 KB
-
Native-structure-based-modeling-and-simulation-of-biomolecular-systems-per-mouse-click-12859 2014 6561 MOESM1 ESM.ogv 1 min 56 s, 1,024 × 1,024; 7.32 MB
-
Native-structure-based-modeling-and-simulation-of-biomolecular-systems-per-mouse-click-12859 2014 6561 MOESM2 ESM.ogv 2 min 10 s, 1,024 × 1,024; 8.92 MB
-
Native-structure-based-modeling-and-simulation-of-biomolecular-systems-per-mouse-click-12859 2014 6561 MOESM3 ESM.ogv 1 min 36 s, 1,024 × 1,024; 7.94 MB
-
Neck nanowire.webm 2 min 11 s, 484 × 444; 5.32 MB
-
Ni nanorod tension simulation MD.png 719 × 1,570; 629 KB
-
Operating-Mechanism-and-Molecular-Dynamics-of-Pheromone-Binding-Protein-ASP1-as-Influenced-by-pH-pone.0110565.s001.ogv 2 min 13 s, 660 × 568; 64.74 MB
-
Operating-Mechanism-and-Molecular-Dynamics-of-Pheromone-Binding-Protein-ASP1-as-Influenced-by-pH-pone.0110565.s002.ogv 2 min 13 s, 712 × 584; 83.46 MB
-
-
-
P53-dynamics-upon-response-element-recognition-explored-by-molecular-simulations-srep17107-s2.ogv 40 s, 512 × 512; 9.16 MB
-
P53-dynamics-upon-response-element-recognition-explored-by-molecular-simulations-srep17107-s3.ogv 53 s, 512 × 512; 6.52 MB
-
Particle Simulation Over a Flat Plate.gif 512 × 512; 5.19 MB
-
Photoswitching-Free-FRAP-Analysis-with-a-Genetically-Encoded-Fluorescent-Tag-pone.0107730.s001.ogv 24 s, 300 × 300; 4.05 MB
-
Photoswitching-Free-FRAP-Analysis-with-a-Genetically-Encoded-Fluorescent-Tag-pone.0107730.s002.ogv 24 s, 300 × 300; 14.65 MB
-
-
-
-
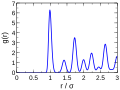
Radial Distribution Function of Liquid Argon.png 1,808 × 1,923; 52 KB
-
Radial Distribution Function of Solid Argon.svg 283 × 213; 27 KB
-
Radial distribution functions of liquid argon and water.png 1,781 × 1,768; 66 KB
-
-
RDF molekula vode.png 861 × 614; 6 KB
-
RdRp Graphical abstract.jpg 3,581 × 2,651; 3.35 MB
-
Role-of-Histone-Tails-in-Structural-Stability-of-the-Nucleosome-pcbi.1002279.s010.ogv 20 s, 720 × 576; 4.06 MB
-
Role-of-Histone-Tails-in-Structural-Stability-of-the-Nucleosome-pcbi.1002279.s011.ogv 0.0 s, 1,280 × 736; 5.26 MB
-
Role-of-Histone-Tails-in-Structural-Stability-of-the-Nucleosome-pcbi.1002279.s012.ogv 9.9 s, 720 × 576; 3.01 MB
-
Role-of-Histone-Tails-in-Structural-Stability-of-the-Nucleosome-pcbi.1002279.s013.ogv 20 s, 720 × 576; 2.83 MB
-
-
Schematic of a replica exchange molecular dynamics simulation.svg 363 × 155; 75 KB
-