Category:Protein structures
Jump to navigation
Jump to search
three-dimensional arrangement of atoms in an amino acid-chain molecule | |||||
| Upload media | |||||
| Subclass of |
| ||||
|---|---|---|---|---|---|
| |||||
Subcategories
This category has the following 25 subcategories, out of 25 total.
*
A
B
- BBSome (4 F)
C
D
F
H
M
N
- NMR structures of proteins (30 F)
P
- Protein structures from PDB (38556 F)
- Protein structural homology (10 F)
Q
R
- RFdiffusion (15 F)
S
T
V
Media in category "Protein structures"
The following 200 files are in this category, out of 1,060 total.
(previous page) (next page)-
1-s2.0-S0166354209005385-gr10.jpg 386 × 325; 64 KB
-
14-3-3 sigma in complex with TAZ pS89 peptide - Protein Data Bank.png 654 × 967; 363 KB
-
153-PyruvateDehydrogenaseComplex pyruvatedehydrogenase.tif 2,572 × 2,788; 20.55 MB
-
1a02.png 500 × 500; 147 KB
-
1ATN - Actin monomer subdomains.png 644 × 584; 383 KB
-
1CEX.jpg 500 × 500; 51 KB
-
1hl5.jpg 1,000 × 797; 238 KB
-
1k5m.jpg 1,000 × 797; 414 KB
-
1n25.jpg 1,000 × 797; 341 KB
-
1tf7.jpg 1,000 × 794; 340 KB
-
1tt0.jpg 1,000 × 794; 361 KB
-
2,4 Hexadienoyl-CoA with DECR1.png 640 × 434; 52 KB
-
202404 human deoxyhemoglobin.svg 400 × 400; 220 KB
-
202404 human oxyhemoglobin.svg 400 × 400; 189 KB
-
2gpq asym r 500.jpg 500 × 500; 19 KB
-
2KM6NLRP7NMR.png 970 × 832; 1.32 MB
-
2NBIAZOOMEDOUT.jpg 468 × 315; 23 KB
-
2o28.png 2,400 × 2,400; 1.33 MB
-
382018 3Q-1R zero ionic layer structure on Pymol.png 573 × 549; 228 KB
-
3D structure AsKC11.png 165 × 298; 24 KB
-
3D Structure of Legumin Protein.png 676 × 648; 301 KB
-
3D structure of napin.png 599 × 532; 209 KB
-
3D Structure of the U24-ctenitoxin-Pn1a protein.png 1,242 × 730; 463 KB
-
3D Structure SBK3.jpg 200 × 200; 19 KB
-
3d tRNA.png 1,061 × 1,056; 525 KB
-
3F73.png 864 × 890; 363 KB
-
3Q-1R zero ionic layer structure labeled.jpg 573 × 549; 206 KB
-
3TSS PDB.png 2,559 × 1,855; 828 KB
-
3X2R.png 833 × 891; 453 KB
-
4A0L CUL4B.png 360 × 295; 65 KB
-
4REY side helix.png 413 × 395; 225 KB
-
5-HT1A receptor.jpg 578 × 785; 136 KB
-
5-HT1D receptor.jpg 578 × 785; 142 KB
-
5-HT1E receptor.jpg 578 × 785; 132 KB
-
5-HT1F receptor.jpg 578 × 785; 139 KB
-
5AR structure.png 970 × 584; 493 KB
-
670 crystal.jpg 452 × 603; 153 KB
-
7E5N TROP2extracellular dimer.png 2,117 × 2,160; 1.44 MB
-
A1-дев'ять.png 400 × 615; 16 KB
-
A1-один.png 400 × 615; 15 KB
-
A2-два.png 400 × 615; 49 KB
-
A2-десять.png 400 × 615; 50 KB
-
A3-одинадцять.png 400 × 615; 18 KB
-
A3-три.png 400 × 615; 22 KB
-
A4-дванадцять.png 400 × 615; 51 KB
-
A4-чотири.png 400 × 615; 52 KB
-
A5-п'ять.png 400 × 615; 15 KB
-
A5-тринадцять.png 400 × 615; 16 KB
-
A6-чотирнадцять.png 400 × 615; 12 KB
-
A6-шість.png 400 × 615; 13 KB
-
A7-п'ятнадцять.png 400 × 615; 18 KB
-
A7-сім.png 400 × 615; 22 KB
-
A8-вісім.png 400 × 615; 16 KB
-
A8-шістнадцять.png 400 × 615; 15 KB
-
Aa structure function (2).svg 284 × 308; 753 bytes
-
Aa structure function.svg 286 × 312; 5 KB
-
AAL67520.1 1..606.pdf 1,891 × 460; 20 KB
-
Abc domain.jpg 3,965 × 2,895; 676 KB
-
Abc-sav.jpg 2,150 × 1,943; 312 KB
-
ABCC11 protein structure scheme 01.svg 1,294 × 700; 330 KB
-
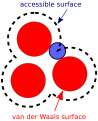
Accessible surface.svg 587 × 720; 9 KB
-
AcetaldehydeDehydrogenase-1NVM.png 786 × 545; 202 KB
-
Acetylene Hydratase (PDB code 2E7Z).png 1,450 × 1,061; 891 KB
-
Actin Filament.jpg 189 × 246; 13 KB
-
Activin inhibin.png 1,403 × 521; 25 KB
-
ADAR Protein 2.png 1,000 × 940; 568 KB
-
ADAR Protein 3.png 800 × 940; 499 KB
-
ADAR Protein.png 1,120 × 940; 564 KB
-
ADAR2 deaminase domain RNA co-crystal PDB 5ED1.png 1,500 × 1,500; 933 KB
-
Adeno-associated virus serotype AAV2.jpg 1,555 × 1,450; 729 KB
-
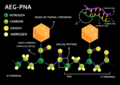
AEG-PNA.png 2,800 × 1,980; 1,003 KB
-
AF-A0A167FEV7-F1.stl 5,120 × 2,880; 44.99 MB
-
AF-A0A2V0PLL6-F1 (1).png 513 × 569; 117 KB
-
AF-A0A2V0PLL6-F1 (2).png 605 × 542; 116 KB
-
AF-A0A2V0PLL6-F1 (3).png 594 × 466; 121 KB
-
AF-E9Q7G0-F1.png 501 × 355; 85 KB
-
AF-P59853-F1.png 547 × 224; 50 KB
-
AF-Q63HQ2-F1.png 1,127 × 715; 344 KB
-
AF-Q9ULY5-F1.png 390 × 410; 47 KB
-
AhR-Hsp90-XAP2 complex PDB 7ZUB.png 1,344 × 1,000; 1.01 MB
-
AIM2PYDside.png 780 × 850; 482 KB
-
AIM2sidetopdown.png 1,612 × 832; 1.28 MB
-
Alignment of thioredoxins.png 629 × 538; 53 KB
-
Alpha Arrestin Structure.jpg 1,830 × 1,780; 213 KB
-
Alpha beta structure (1).png 815 × 438; 104 KB
-
Alpha beta structure (2).png 815 × 438; 98 KB
-
Alpha beta structure (full).png 815 × 438; 116 KB
-
Alpha Fold Tertiary c4orf54 structure.png 1,706 × 964; 1.03 MB
-
Alpha solenoid pp2a 2iae with single repeat center.png 1,024 × 1,024; 665 KB
-
Alpha-h2.png 162 × 435; 2 KB
-
Alpha-h3.png 162 × 435; 4 KB
-
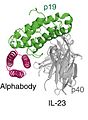
AlphabodyXRC.jpg 241 × 330; 34 KB
-
AlphaHelixProtein.jpg 197 × 524; 58 KB
-
AMH & AMHR2.png 2,000 × 2,000; 1,006 KB
-
Amido (1).svg 744 × 1,052; 20 KB
-
Amminoacidi e proteine.png 1,007 × 709; 143 KB
-
Anaglyph image of a protein DHFR.png 1,120 × 887; 655 KB
-
Animación-proteína-CDC13.gif 598 × 738; 5.51 MB
-
ANKRD35 protein structure.jpg 1,461 × 735; 82 KB
-
Annotated 3D AMHR2 protein showing key motifs.png 1,244 × 926; 251 KB
-
Annotated BEND2 I-TASSER.png 808 × 662; 243 KB
-
Annotated Tertiary Structure of Isoform X1.png 672 × 400; 107 KB
-
AnPRT structure.png 640 × 480; 178 KB
-
Antibody Structure.png 602 × 292; 5 KB
-
APT tunnel.png 1,218 × 2,208; 2.57 MB
-
ARN pol II en accion.jpg 904 × 580; 36 KB
-
Arrangement of subunits and domains within the BBSome core.png 630 × 540; 546 KB
-
Aryl hydrocarbon receptor PDB 7ZUB.png 1,344 × 1,000; 564 KB
-
Asdadwgtfw3w.jpg 375 × 319; 16 KB
-
Basic Protein Structure.png 1,432 × 634; 148 KB
-
BAT5 protein structure.jpg 500 × 350; 33 KB
-
BBDEP Newman diagram.png 7,447 × 6,050; 1.56 MB
-
BBE-like enzyme structure.jpg 620 × 354; 52 KB
-
Bcl2.png 1,362 × 681; 379 KB
-
BDNF NT4.png 5,000 × 6,600; 10.46 MB
-
Beta-catenin-in-cancer.png 776 × 744; 43 KB
-
BI tert structure.jpg 500 × 150; 31 KB
-
Bifunctional ppGpp synthase hydrolase SpoT.png 802 × 920; 382 KB
-
BLIP-Bla complex.png 639 × 670; 209 KB
-
Bovine Papillomavirus Capsid.png 1,066 × 1,066; 1.97 MB
-
Bronsted character of ionizing groups in proteins.png 571 × 818; 74 KB
-
C11orf98 teritary.png 722 × 330; 31 KB
-
C13orf42 structure predicted by I-Tasser and annotated in Chimera.png 2,736 × 1,534; 1.53 MB
-
C16orf96 Protein Structure.png 552 × 174; 30 KB
-
C19orf47 tertiary structure from iCn3D.png 876 × 828; 143 KB
-
C19orf67 3D.png 640 × 480; 81 KB
-
C19orf67 Post-Translational Modifications.png 1,156 × 586; 26 KB
-
C4orf50 Alphafold.png 716 × 350; 171 KB
-
C4orf50 i-TASSER.png 700 × 688; 177 KB
-
C4orf50 phyre.png 326 × 158; 53 KB
-
C5orf36 Predicted Tertiary Structure.png 640 × 434; 70 KB
-
C6orf58 Protein Structure.jpg 713 × 340; 35 KB
-
C6orf58 protein structure.png 1,103 × 544; 40 KB
-
CADENES NOM.jpg 425 × 292; 29 KB
-
Calcium Binding to Citrin.png 2,374 × 2,079; 3.71 MB
-
Calmodulin Binding sites.gif 615 × 544; 75 KB
-
CalmodulinPYMOL.jpg 2,048 × 2,008; 499 KB
-
Captura de pantalla 2021-11-09 141336.png 934 × 586; 135 KB
-
Captura de pantalla 2023-10-30 a las 17.09.44 copia.png 1,140 × 594; 111 KB
-
Carboxipeptidasa B estructura.jpg 1,392 × 1,940; 929 KB
-
Carboxipeptidasa B.jpg 1,409 × 1,984; 943 KB
-
CARD11.png 868 × 614; 239 KB
-
CARD11.tiff 774 × 442; 1.31 MB
-
Carnobacteriocin B2.png 500 × 500; 65 KB
-
CATH hierarchy.png 2,502 × 2,992; 1.46 MB
-
CAVE protein structure.jpg 4,256 × 2,832; 5.7 MB
-
Cavin 1.png 1,081 × 702; 86 KB
-
CBlast 72-272 asparagines.png 326 × 452; 118 KB
-
CCDC177 Tertiary Structure.png 1,440 × 1,186; 421 KB
-
CCDC188 Homodimer.png 684 × 680; 149 KB
-
CCDC60 Structure.png 1,200 × 1,200; 343 KB
-
CCN1 Protein Structure.jpg 960 × 720; 38 KB
-
CCN1 Protein Structure.svg 534 × 430; 42 KB
-
CCR7 receptor.png 1,800 × 1,788; 692 KB
-
CDO full structure.png 3,057 × 1,523; 576 KB
-
CFTR protein structure scheme 02.svg 980 × 680; 127 KB
-
Charge of TMEM211.png 512 × 310; 38 KB
-
Chignolin.jpg 680 × 563; 19 KB
-
Chimera render of Smad4 protein from PDB 5MEZ.png 2,000 × 1,243; 773 KB
-
Citrin Structure.png 589 × 379; 138 KB
-
Citrine proteine.tif 1,872 × 1,691; 11.42 MB
-
ClpP structure.png 605 × 495; 322 KB
-
Codeinone reductase.png 1,700 × 755; 307 KB
-
Coiled coil structure.gif 1,650 × 1,682; 281 KB
-
COMPARACIÓN.png 1,074 × 784; 250 KB
-
Conserved residues.svg 193 × 500; 102 KB
-
Convergence Thr.png 2,174 × 634; 516 KB
-
Cover (15276238426).jpg 1,200 × 660; 787 KB
-
CPSF2 highlighted within CPSF complex.png 766 × 589; 343 KB
-
CRELD2 Structure.png 514 × 380; 64 KB
-
Crmaedit.png 667 × 681; 159 KB
-
Cryoem groel.jpg 1,024 × 1,024; 499 KB
-
Cryoem groel.png 1,024 × 1,024; 609 KB
-
Crystal structure of Cry6Aa toxin.gif 567 × 446; 58 KB
-
Crystal structure of E. coli Exodeoxyribonuclease I.png 3,000 × 2,100; 1.76 MB
-
Crystal Structure of N-Type Channel.png 350 × 350; 76 KB
-
Crystal Structure of Prp8.png 569 × 459; 164 KB
-
Crystal structure of Synechocystis ACO.png 2,950 × 2,450; 1.39 MB
-
Cubilin-FI.png 5,025 × 4,616; 4.93 MB
-
Cubilina.png 2,941 × 3,106; 3.84 MB
-
CUBN-AMN 2.gif 1,676 × 1,396; 11.47 MB
-
CUL4A-brown-2HYE.png 408 × 254; 57 KB
-
Cumulative Unique Membrane Protein Structures by Year.png 3,500 × 2,000; 102 KB
-
Current-model-of-TXNDC5-interactions-with-different-compounds.jpg 790 × 428; 78 KB
-
Cut 1UVB.png 1,500 × 1,580; 416 KB
-
CXorf38 Space-filling Model.png 1,101 × 397; 213 KB
-
Cyclic peptide ipglycermide.png 1,250 × 1,017; 912 KB
-
CYP4F2 AF AFP78329F1.png 1,280 × 1,031; 796 KB
-
Cys193closeshot.jpg 468 × 202; 24 KB
-
Cytochrome b5 reductase structure.png 958 × 598; 615 KB
-
DB00031.png 500 × 500; 179 KB
-
DHOQO structure colored by domains.png 782 × 586; 267 KB
-
Dibujo DK y alfaC.png 724 × 652; 368 KB
-
DIRAS1 Protein Structure.jpg 500 × 500; 45 KB