Category:Post-translational modifications
Jump to navigation
Jump to search
covalent and generally enzymatic modification of proteins during or after protein biosynthesis | |||||
| Upload media | |||||
| Instance of | |||||
|---|---|---|---|---|---|
| Subclass of |
| ||||
| Different from | |||||
| |||||
Subcategories
This category has the following 9 subcategories, out of 9 total.
Media in category "Post-translational modifications"
The following 159 files are in this category, out of 159 total.
-
-
ADAMTS3 proteolytic processing of Reelin.webp 1,939 × 1,052; 79 KB
-
AICAR transformylase active site.png 776 × 623; 311 KB
-
AICAR transformylase mechanism.png 3,787 × 922; 87 KB
-
-
-
Allatostatin-A-Signalling-in-Drosophila-Regulates-Feeding-and-Sleep-and-Is-Modulated-by-PDF-pgen.1006346.s023.ogv 3 min 40 s, 854 × 480; 6.47 MB
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
Basic N-glycan.svg 512 × 887; 33 KB
-
Biosynthesis of N-linked oligosaccharides.svg 1,200 × 600; 148 KB
-
C-mannosylation process.svg 512 × 566; 28 KB
-
C16orf82 Diagram With PTM.png 309 × 40; 5 KB
-
C17orf75 protein schematic.png 1,076 × 546; 24 KB
-
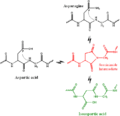
Citrullination.svg 900 × 510; 19 KB
-
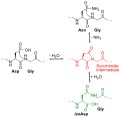
Deamidation Asn Gly.png 699 × 678; 14 KB
-
Deamidation Asn Gly.svg 512 × 501; 38 KB
-
Diubiquitin-lysine-48.png 1,100 × 750; 282 KB
-
Diubiquitin-lysine-63.png 1,100 × 750; 197 KB
-
Domain structure of mTOR.png 472 × 130; 9 KB
-
Entamoeba-histolytica-Cysteine-Proteinase-5-Evokes-Mucin-Exocytosis-from-Colonic-Goblet-Cells-via-ppat.1005579.s004.ogv 4.9 s, 1,104 × 1,104; 6.74 MB
-
Entamoeba-histolytica-Cysteine-Proteinase-5-Evokes-Mucin-Exocytosis-from-Colonic-Goblet-Cells-via-ppat.1005579.s005.ogv 4.5 s, 1,236 × 1,236; 4.43 MB
-
-
-
Figure4trd.png 897 × 562; 58 KB
-
Formation of 5-oxoproline from N-terminal Gln.svg 512 × 143; 9 KB
-
GeneticCode22.svg 725 × 950; 303 KB
-
Glutamine synthase regulation by PII proteins.png 1,291 × 913; 233 KB
-
GPI.pdf 1,500 × 1,125; 15 KB
-
Hypoxia-Regulates-mTORC1-Mediated-Keratinocyte-Motility-and-Migration-via-the-AMPK-Pathway-pone.0169155.s004.ogv 8.7 s, 1,530 × 648; 2.42 MB
-
Hypoxia-Regulates-mTORC1-Mediated-Keratinocyte-Motility-and-Migration-via-the-AMPK-Pathway-pone.0169155.s005.ogv 8.7 s, 1,020 × 649; 1.73 MB
-
Insulin path.svg 1,488 × 2,105; 282 KB
-
Insulinpath.png 1,488 × 2,105; 353 KB
-
K63-Linked-Ubiquitination-Targets-Toxoplasma-gondii-for-Endo-lysosomal-Destruction-in-IFNγ-ppat.1006027.s001.ogv 3.4 s, 1,024 × 1,024; 307 KB
-
K63-Linked-Ubiquitination-Targets-Toxoplasma-gondii-for-Endo-lysosomal-Destruction-in-IFNγ-ppat.1006027.s002.ogv 2.8 s, 1,024 × 1,024; 134 KB
-
-
K63-Linked-Ubiquitination-Targets-Toxoplasma-gondii-for-Endo-lysosomal-Destruction-in-IFNγ-ppat.1006027.s005.ogv 3.3 s, 1,772 × 1,099; 1.85 MB
-
-
-
LOC101059915 Diagram.png 860 × 217; 20 KB
-
Lysine 5 hydroxy.png 149 × 252; 2 KB
-
Lysine Tyrosylquinone Mediated Oxn of Lysine.svg 685 × 189; 21 KB
-
Methyl arginine.tif 519 × 206; 313 KB
-
Methyl lysine.tif 450 × 165; 218 KB
-
Methylglutamate.png 155 × 241; 2 KB
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
Myristoylation.pdf 1,500 × 1,125; 22 KB
-
N6,N6-dimethyllysine.svg 375 × 470; 8 KB
-
N6-methyllysine.svg 375 × 470; 8 KB
-
Nisin RiPP.png 3,703 × 2,084; 396 KB
-
Notch signaling pathway reactome.png 3,555 × 1,706; 290 KB
-
Nucleosome Complex model.ogv 8.4 s, 1,280 × 624; 2.31 MB
-
O-GlcNAc clear red.png 903 × 522; 49 KB
-
O-GlcNAc red.png 688 × 375; 42 KB
-
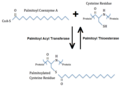
Palmitoylation.png 702 × 509; 68 KB
-
PalmitoylationAnkGc pl.jpg 340 × 360; 76 KB
-
PARP1 with DNA.png 950 × 656; 201 KB
-
Pauci Intro crop.jpg 5,310 × 2,459; 1.52 MB
-
Pauci Intro.jpg 5,999 × 4,498; 1.66 MB
-
PCP complex.png 1,547 × 1,547; 1.4 MB
-
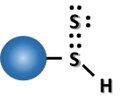
Persulfidation.tif 135 × 112; 80 KB
-
Phosphoserine.tif 219 × 113; 73 KB
-
PhosphoSitePlus TUBA1A.png 2,000 × 1,000; 150 KB
-
Phosphotyrosine.tif 281 × 132; 109 KB
-
Phosporylation of serine residue after.png 1,920 × 976; 192 KB
-
Phosporylation of serine residue before.png 1,920 × 976; 165 KB
-
Plantazolicin RiPP.png 3,657 × 1,032; 228 KB
-
Post-transcriptional modification of pre-mRNA.png 1,316 × 898; 93 KB
-
Post-translational modification by cleavage.png 1,434 × 528; 93 KB
-
Post-translational modification sites of c4orf21.png 980 × 208; 28 KB
-
Post-translational modification through the addition of small chemical groups.png 1,804 × 1,252; 431 KB
-
Post-Translational Modifications.png 745 × 348; 18 KB
-
Posttranslational modification of human CK1δ.jpg 1,386 × 340; 97 KB
-
Pre-NOTCH 72 oicr.png 800 × 1,384; 666 KB
-
Protein adenylylation.png 2,372 × 755; 68 KB
-
Protein-Translation-Enzyme-lysyl-tRNA-Synthetase-Presents-a-New-Target-for-Drug-Development-against-pntd.0005084.s003.ogv 8.1 s, 4,420 × 2,698; 16.54 MB
-
PTMofLysine.svg 496 × 195; 16 KB
-
PZN-biosynthesis.png 7,675 × 6,950; 523 KB
-
-
-
-
-
Shh processing.png 791 × 307; 23 KB
-
Shh processing.svg 790 × 305; 9 KB
-
Summary of Post-Translational Modifications of FAM46B.png 1,365 × 768; 192 KB
-
Tau dominis.jpg 960 × 598; 32 KB
-
Tetraubiquitin-K48-3ALB.png 1,128 × 1,192; 576 KB
-
Tetraubiquitin-K63-3HM3.png 2,552 × 1,544; 601 KB
-
-
-
-
-
-
-
-
-
-
-
-
TMEM248 PTM diagram.png 1,480 × 482; 212 KB
-
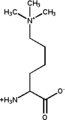
Trimethyllysine.png 148 × 273; 3 KB
-
Trimethyllysine.svg 375 × 500; 9 KB
-
Trunkamide RiPP.png 1,136 × 1,613; 117 KB
-
-
-
-
-
-
-
-
Ubiquitin cartoon-2-.png 900 × 800; 157 KB
-
Ubiquitin cartoon.png 828 × 575; 153 KB
-
Ubiquitin spheres.png 800 × 645; 241 KB
-
Ubiquitin surface.png 942 × 673; 395 KB
-
-
-
-
-
-
-
Ubiquitinylierung.JPG 268 × 489; 13 KB