User talk:Nghaemim
|
Our first steps tour and our frequently asked questions will help you a lot after registration. They explain how to customize the interface (for example the language), how to upload files and our basic licensing policy (Wikimedia Commons only accepts free content). You don't need technical skills in order to contribute here. Be bold when contributing and assume good faith when interacting with others. This is a wiki. More information is available at the community portal. You may ask questions at the help desk, village pump or on IRC channel #wikimedia-commons (webchat). You can also contact an administrator on their talk page. If you have a specific copyright question, ask at the copyright village pump. |
|
-- 00:47, 30 October 2011 (UTC)
DNA Replicases from a Bacterial Perspective
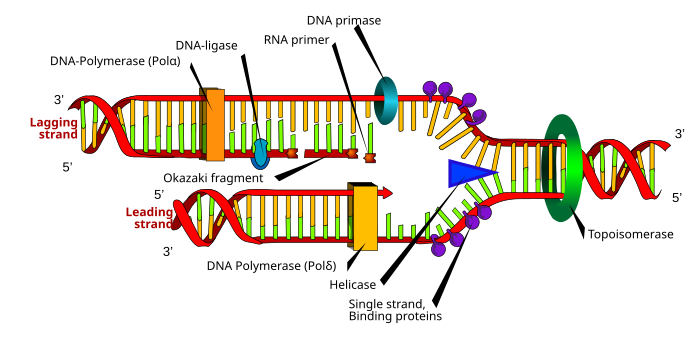
[edit]DNA is replicated through the interaction of the template strand with a huge protein complex called the replication complex, which catalyze the reaction involved. All chromosomes have at least one base sequence, called the origin of replication (ori), to which this reapplication complex initially binds. This binding is based on the recognition of the different nucleotide bases by proteins. DNA replicates in both directions from the origin of replication, forming two replication forks. Both of the separated strands of the parent molecule act as templates simultaneously, and the formation of the new strands is guided by complementary base pairing. Replication begins at specific “origin of replication” sequences to which this replication complex initially binds, and forms replication “bubbles”. Two important rules for DNA synthesis are DNA polymerases cannot initiate a strand, and DNA synthesis always proceeds in the 5’ to 3’ direction. DNA needs a complementary short single strand of RNA which is called primer in addition to template in order to be initiated. For the second rule, when nucleotide are added to free hydroxyls, not to 5’ phosphate, we see that the template is actually traversed in the 3’ to 5’ direction, since the two strands are antiparallel. Thus, how can the “lagging” strand be replicated? The answer is discontinuously in other words, by way of Okazaki fragments. In E. coli at least, both strands are synthesized by a single enzyme complex. All replication complexes contain several proteins with different roles in DNA replication. Some of many proteins required for DNA replication in E-coli are being considered as below. An origin binding protein which is called DnaA initiates the process of replication by recognizing and binding to oriC, partially unwinding the helix. A helicase (DnaB protein) is there to continue the unwinding process. Complex needs another protein for releifing the tension and converted to happy way that was before by cutting both of the sugar phosphate back bone and move one of the ends around and then relegate. This protein is called a topoisomerase for “relaxing” the overwound DNA that accumulates ahead of helicase. There is another enzyme (primase) which is an RNA polymerase that lays down short about15 nucleotides lengths of RNA, priming DNA synthesis. Other protein is single stranded binding protein, ssb. Its function is to multiple copies of this relatively simple protein adhere to any single stranded DNA, preventing it from reattaching. Another important protein in the replication complex is a DNA polymerase III “dimmer” (1 monomer for each strand). In E.coli, DNA polymerase III is the primary replicative enzyme. Each monomer is very large, about 600 kilodaltons, complex of 10 subunit polypeptides. The alpha subunit contains the active site for nucleotide addition, and a dimmer of the beta subunit forms a doughnut-shaped ring surrounding one strand of DNA, preventing the polymerase from falling off. This called beta clamp. The epsilon subunit is responsible for proofreading. DNA polymerase I is responsible for removing the RNA primers and replacing them, one nucleotide at a time, with deoxyribonucleotide (DNA). It also functions in various DNA repair mechanisms. Finally, the enzyme DNA ligase is there for joining the Okazaki fragment after DNA polymerase I has acted. There is a model at the top of the page to show DNA replication process. Accuracy and speed in E. coli is about 500 nucleotides strand per second. The error rate of the polymerase III alpha subunit is 1/10000. This would be intolerably high. The proofreading activity of the epsilon subunit corrects most of these errors. A variety of other DNA repair system correct all manner of environmentally-induced mutations, and one of these the MutHLS system in E. coli, also corrects mismatched nucleotides in freshly synthesized DNA. The final correct rates is 1/1000000000 (1 mistake per billion nucleotides added). Numerous systems exist for repairing almost all imaginable types of mutation. In E. coli, over 100 genes are devoted to this protective function.